ASCoVPred is a collection of compounds descriptors / fingerprints (FPs) calculator tool (PaDEL v2.21) and different machine learning (ML) algorithms (implemented in Weka v3.8.2) that are used to build the ML-based quantitative structure-activity relationship (QSAR) prediction models. These QSAR models can be used for ML-assisted prediction of anti-SARS-CoV-2 activity and human cell toxicity of compounds. |
Figure 1. A systematic computational approach used for building ML-based QSAR prediction models and their usage by users. (A). Flow diagram depicting the overall strategy used to train, evaluate and build the ML-based QSAR prediction models. (B). The best prediction models can be used by users to predict the anti-SARS-CoV-2 activity and human cell toxicity of compounds. |
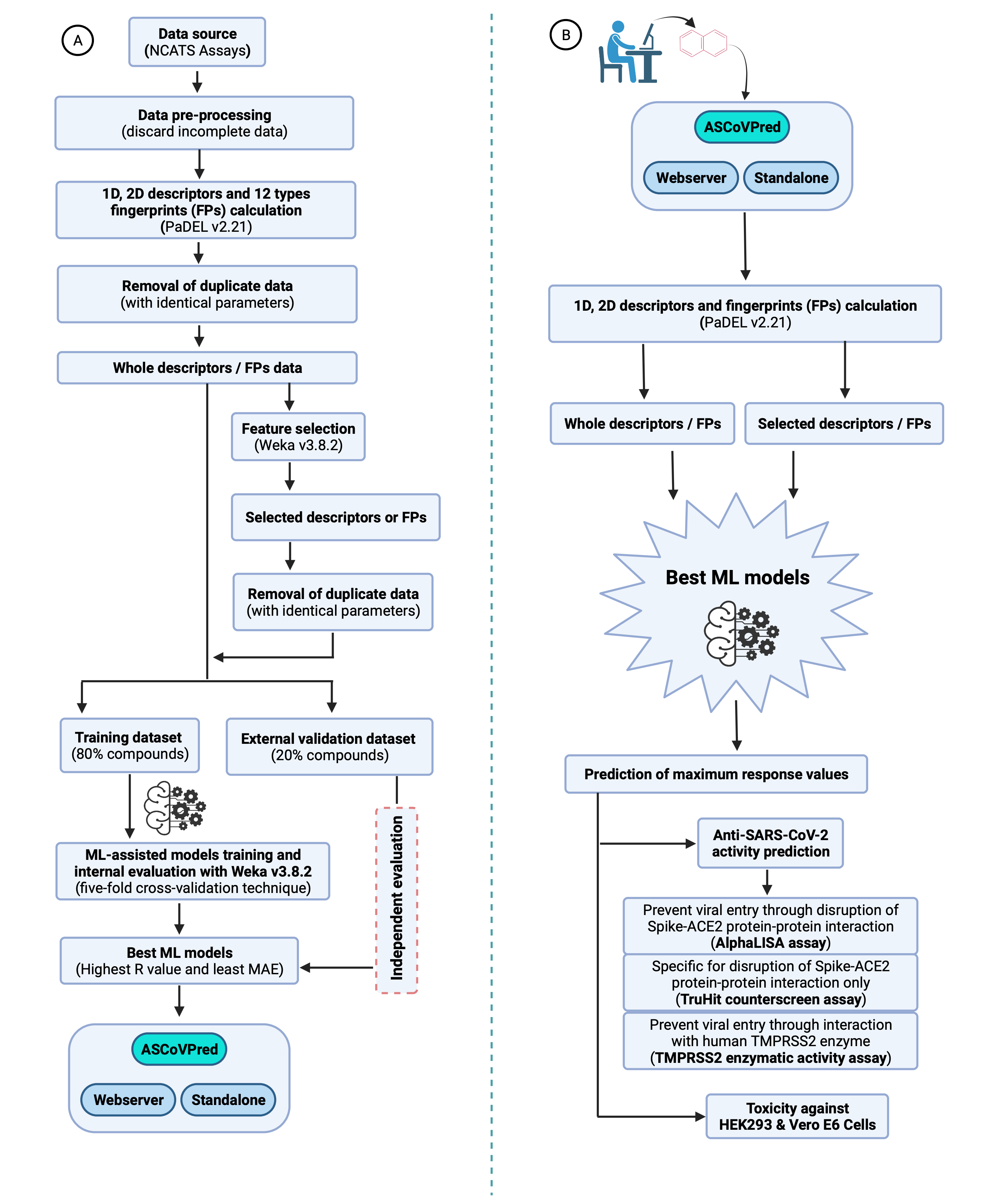 |
Currently, the web server version of ASCoVPred allows users to submit a single compound (at a time) for the prediction. However, the standalone version of the ASCoVPred (tested on Linux and Mac OS), allow its users to screen or process large compound libraries (in batch mode) on their local machines. The standalone version of ASCoVPred (for Linux and Mac OS), for high-throughput screening (HTS) of large chemical libraries, can be downloaded from DOWNLOAD |