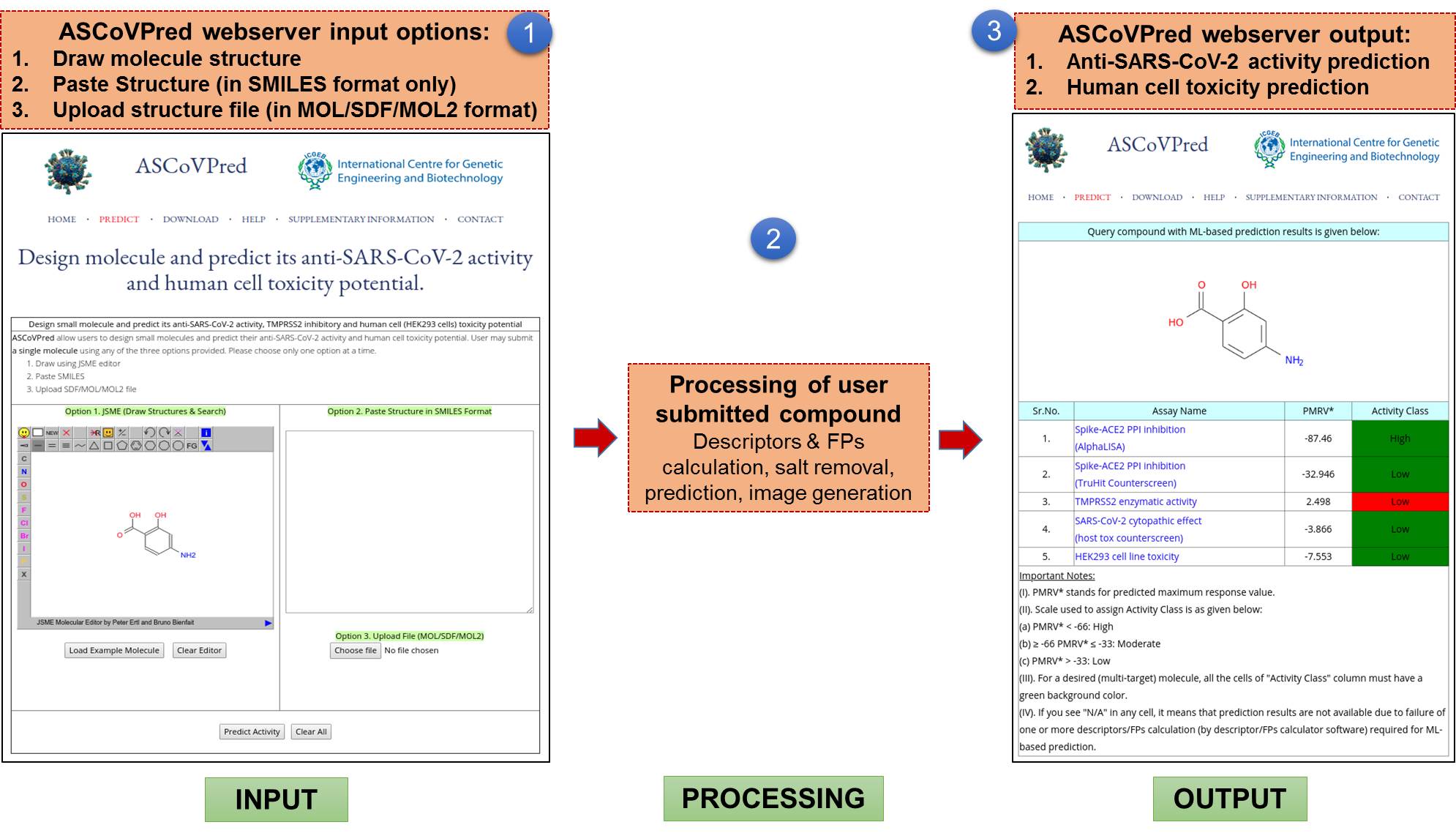
(A). ASCoVPred web server:ASCoVPred provides its users a user-friendly web-interface to utilize the ML-based quantitative structure-activity relationship (QSAR) prediction models. These QSAR models can be used for ML-assisted prediction of anti-SARS-CoV-2 activity and human cell toxicity of compounds. Currently, the web server version of ASCoVPred allows users to submit a single compound (at a time) for the prediction. The Figure 1 shows functioning of ASCoVPred web server. |
|
 |
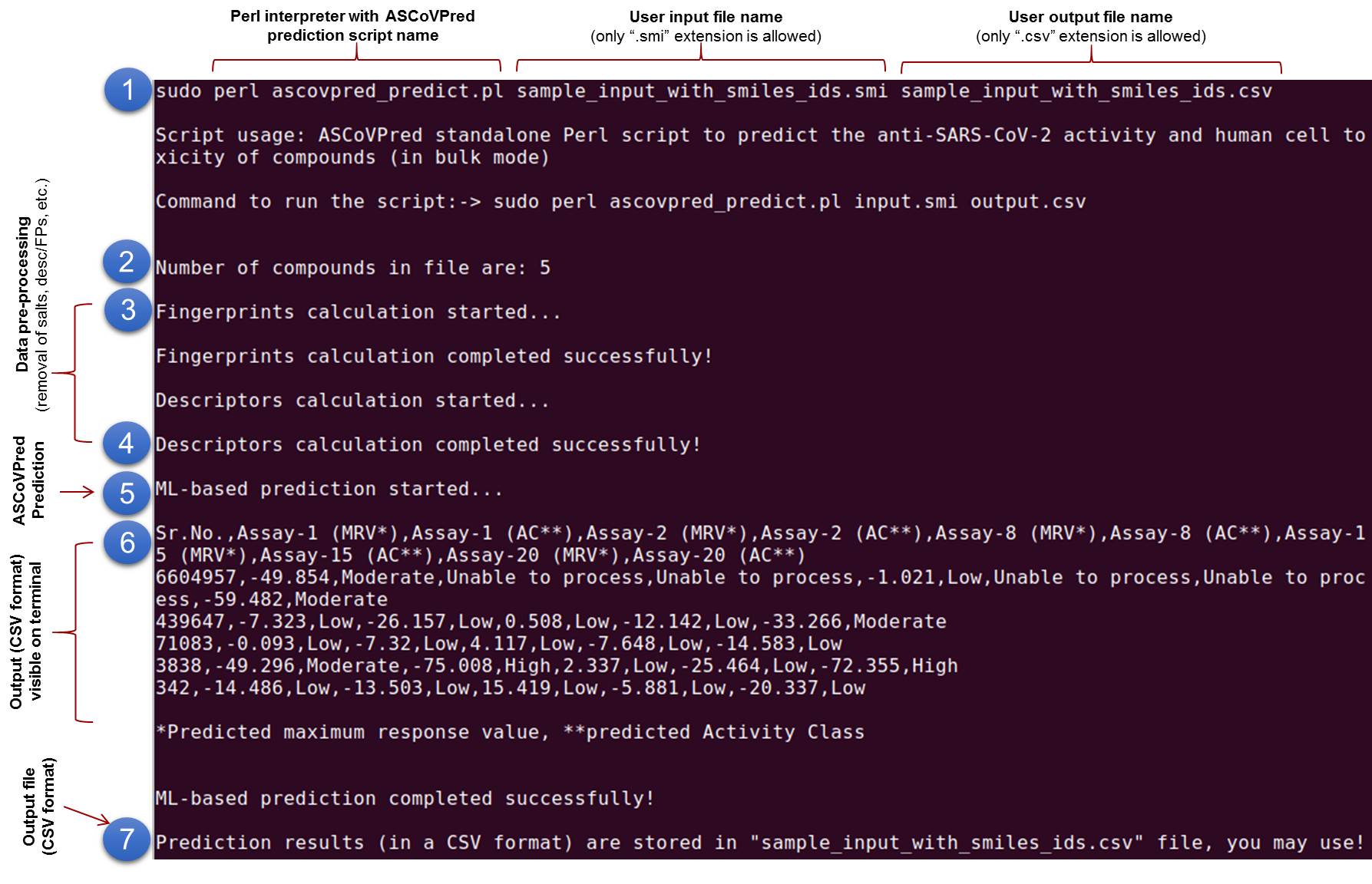
(B). ASCoVPred standalone:The standalone version of the ASCoVPred (tested on Linux and Mac OS), allows its users to screen or process large compound libraries (in batch mode) on their local machines. The standalone version of ASCoVPred (for Linux and Mac OS), for high-throughput screening (HTS) of large chemical libraries, can be downloaded from DOWNLOAD. The functioning of ASCoVPred standalone is shown by Figure 2 given below. |
Figure 2. Functioning of ASCoVPred standalone in action. As depicted in the figure, prediction through ASCoVPred standalone is a multi-step process: (1). Command to run the ASCoVPred prediction Perl script (named "ascovpred_predict.pl"), is typed on the terminal window along with "input file name" (with ".smi" extension only) and "output file name" (with ".csv" extension only) as command-line arguments. (2). Number of compounds (in input file) are counted by the script and displayed on the terminal window. (3-4). Compounds' data pre-processing steps such as removal of salts from structures, standardization of nitro groups, calculation of fingerprints (FPs) and descriptors (1D and 2D only), etc. (for the user submitted compounds) takes place in the backend. During this process, a small pop-up window (from PaDEL descriptor calculator) may appear on your screen. This pop-up window will automatically disappear after descriptors and FPs calculation. (5). After successful calculation of fingerprints (FPs) and descriptors, ML-based prediction starts automatically and compounds maximum response values are predicted with the help of different QSAR models (trained and evaluated with assay-wise experimental data). (6). Prediction results are displyed (in csv format) on the terminal screen as well as stored in an output csv file. (7). The output csv file contains first line as header line while others possess compound IDs (Sr.No.) along with the predicted maximum response values (MRV) and predicted activity class (AC) information for each compound in a separate row. Thus, predicted MRV and AC provides putative anti-SARS-CoV-2 activity and human cell toxicity for compounds in a batch mode. The scales used to assign activity class to compounds (based on predicted maximum response values of the molecules) are always shown at the bottom of each prediction output result page of ASCoVPred web server. |
 |
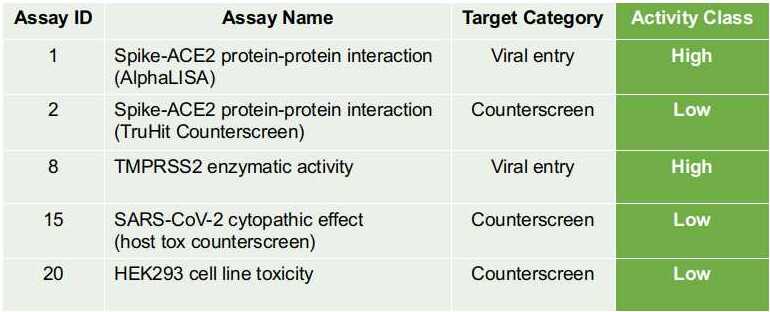
(C). Profile for the desired multi-target molecule on ASCoVPred platform:For any molecule to be an ideal multi-target hit (against both SARS-CoV-2 and human TMPRSS2), it must be predicted as highly active by AlphaLISA assay (Assay ID-1), least active by TruHit counterscreen assay (Assay ID-2), highly active by TMPRSS2 enzymatic activity assay (Assay ID-8), least active by “SARS-CoV-2 cytopathic effect (host tox counterscreen)” assay (Assay ID-15) and HEK293 cell-line assay (Assay ID-20). The desired activity prediction profile for an ideal multi-target molecule is shown in Figure 3. The scales used to assign “Activity Class”, are (also displayed on the ASCoVPred webserver’s prediction results page):(a) PMRV < -66: High (b) ≥ -66 PMRV ≤ -33: Moderate (c) PMRV > -33: Low Where "PMRV" stands for the predicted maximum response value of a query compound. Thus, for a desired (multi-target) molecule, all the cells of "Activity Class" column must have a green background color (Figure 3). |
|
 |