DSP: A database of Disease Susceptibility Genes in Plants
This database is developed to provide the support to plant community in searching and predicting the disease susceptibility genes (DSG). Previously, it has been found that DSG are responsible for making the plant susceptible to various kind of diseases including bacterial, viruses, funagl etc. In this database, users can find the DSG with respect to plant species as well as pathogen type. We have provided simple search and advanced search options where user can search on different parameters. We have also incorporated Blast tool for searching homologs of plant disease susceptibility genes in other crops.
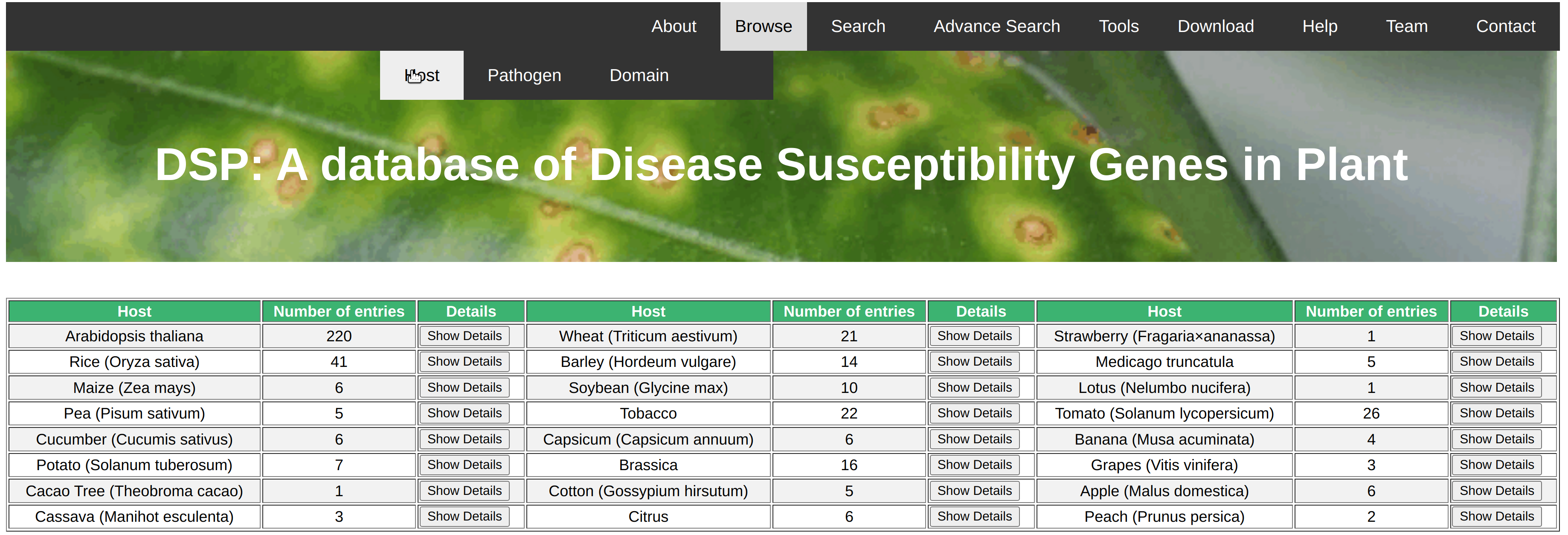
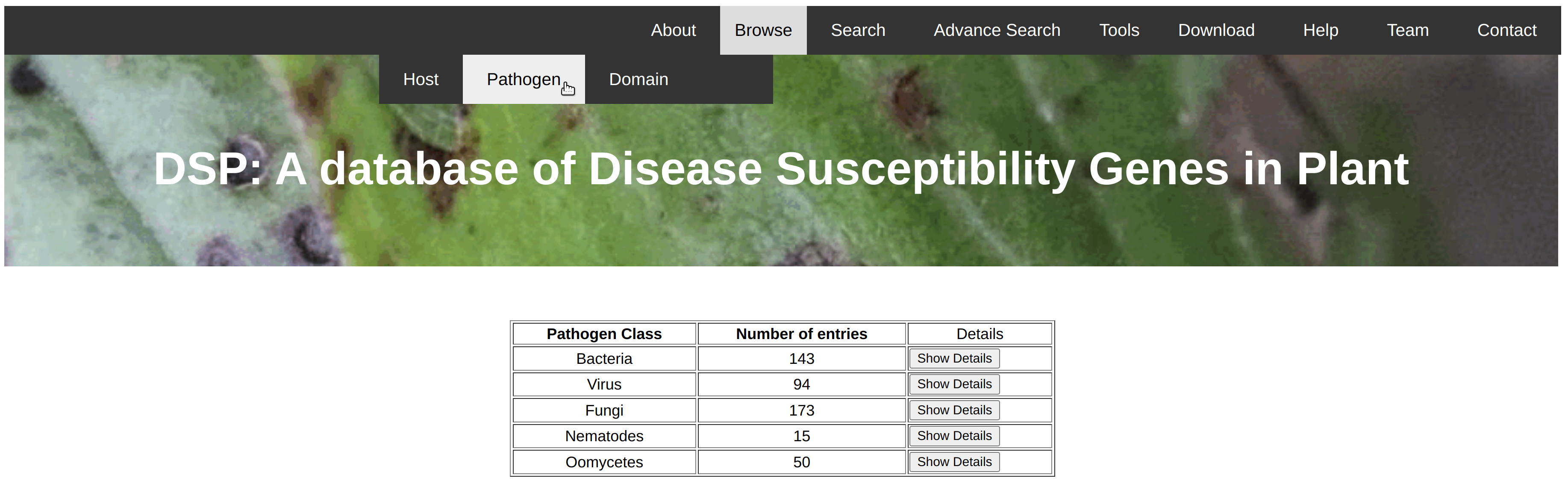
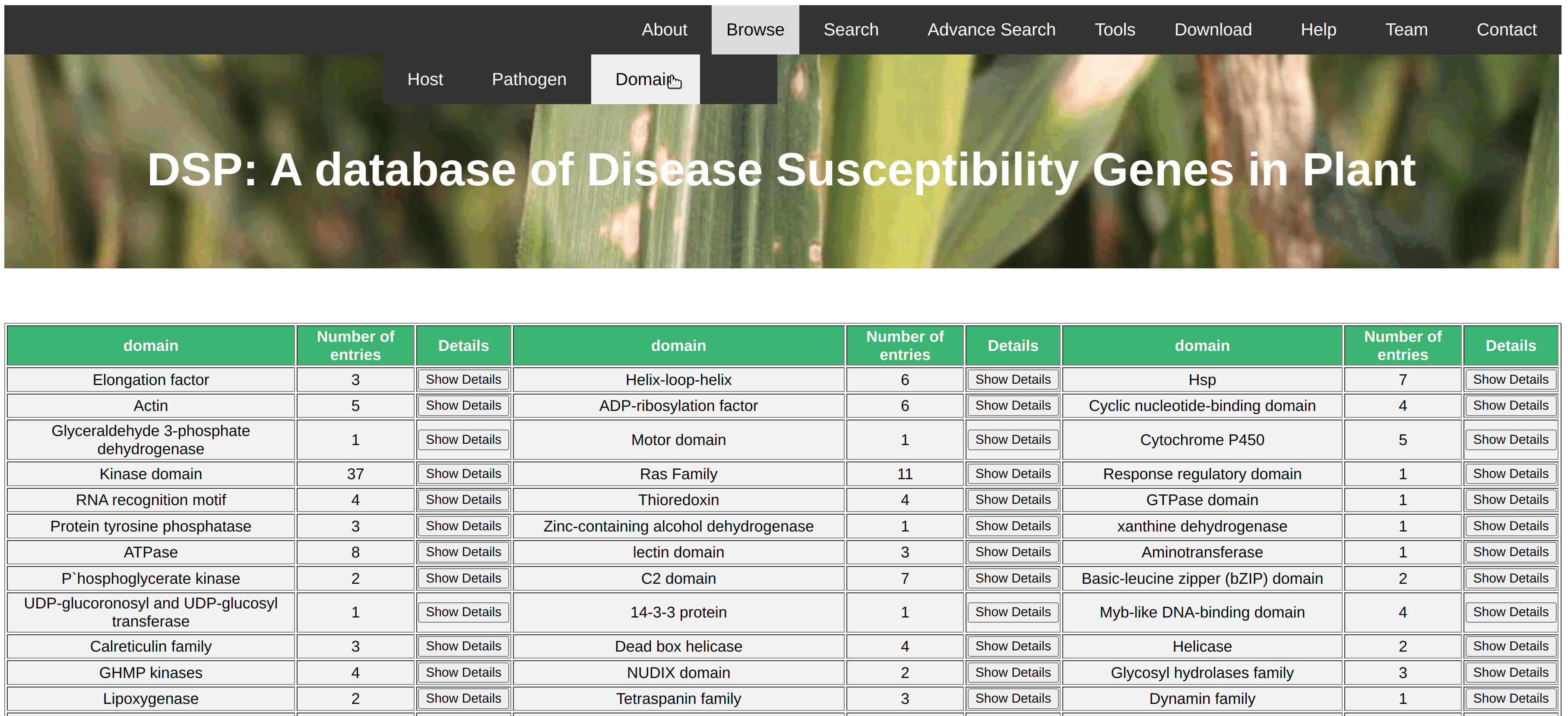
The browse option could be useful to search for all the disease susceptible genes of a particular query like pathogen, host etc. In this database, we have provided 3 browse options for the ease of users. User can browse all the disease susceptible genes based on particular host species like wheat, tomato, rice etc., based on type of pathogens like bacteria, virues, fungi etc., based on protein domains present in th identified disease susceptible genes. The result of query is shown in the same page with no. of genes belongs to the particular class. Show Details button is given along the side of record. After clicking on this information detail of all the respective genes will be shown in the new page in the form of gene name, pathogen name and its class, chromosomal position etc. Further, information about the gene sequence, cds sequence, protein sequence and thier length. no. of exon etc. are given in the detailed page.
Simple search option could be used to search any keyword relatd to S-gene query.
Advance option will be useful for the users with some prior information about the gene or species or pathogens etc. We have provided two options a) exact search b) matching keyword. In the 1st, exact keyword entered by the users will be search in the database. However in the 2nd option, the matching keyword will be search in the database and all the records having complete or a part of keyword matching will be displayed in the result section. Secondly, as shown in Figure-4, user can also restrict your query to a particular column tag so that he/she will get only precise result that could be analyzed further. Users can search the keyword in any of the 15 column or a combination of more than one column or by selecting all the column provided in the search page by clicking on the option of his/her choice.
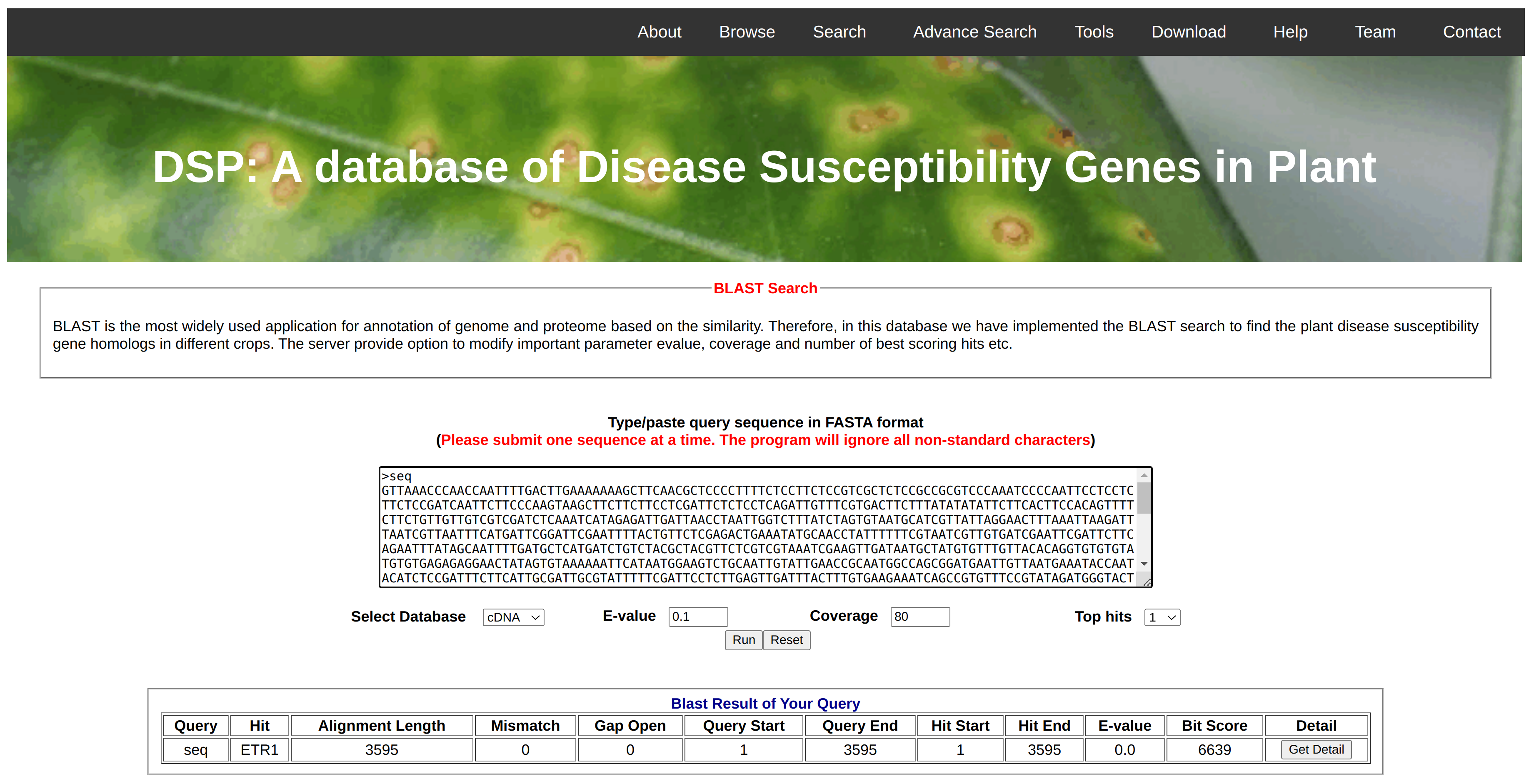
In the tool tab, we have given the Blast search option. This option could be helpful for searching similar genes in other species (orthologs identification) as well as duplicate genes (paralogs identification). User can blast their interest of gene or cds or protein in the local database maintained in the webserver. As shown in window, it only accept the fasta sequence and non-nucleotide character in the DNA/RNA sequence will be ignored. Furhter, user can restrict the results on the basis on e-value, coverage and number of top hit to be shown in the result section. From the results, he/she can check the details of gene matching to his/her query and get an insight about the function and importance of gene.
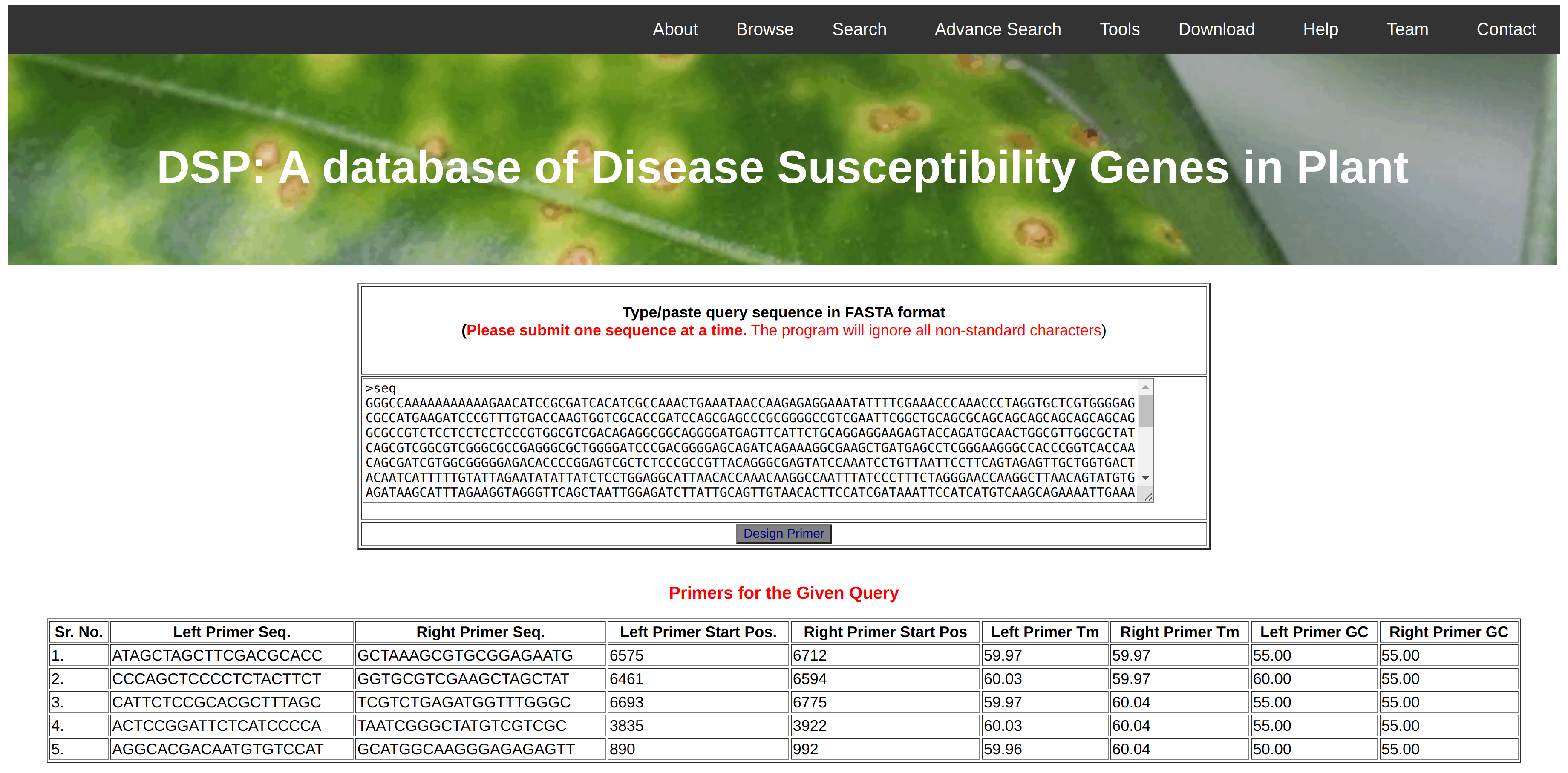
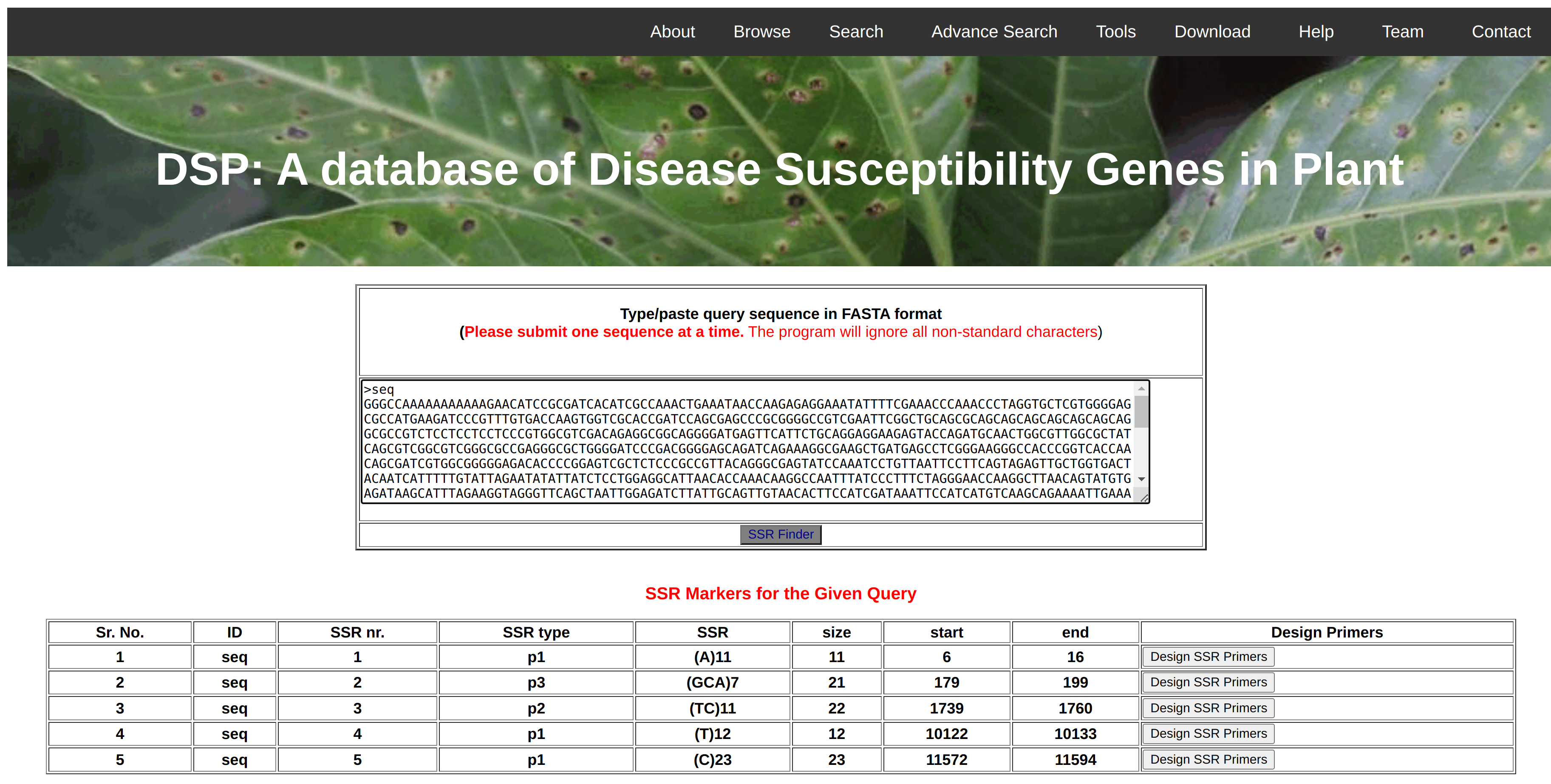
Simple Sequence Repeats (SSR) are the repeats of nucleotides and being used as a markers in the polymorphism studies. In this database, we have implemented the MISA software that will identify the mono, di, tri, tetra, penta, hexa nucleotide repeats as well as complex marker in the given query sequence. Further, the identified markers could be validated in the lab by designing primer for that region. Therefore, we are also providing the facility to design primers for a selected SSR marker using Primer3 Software. Initially, the 250bp upstream and downstream region of the identified marker will be extracted from the main gene sequence and 5 pairs of possible primers could be predicted using the Primer3 software.
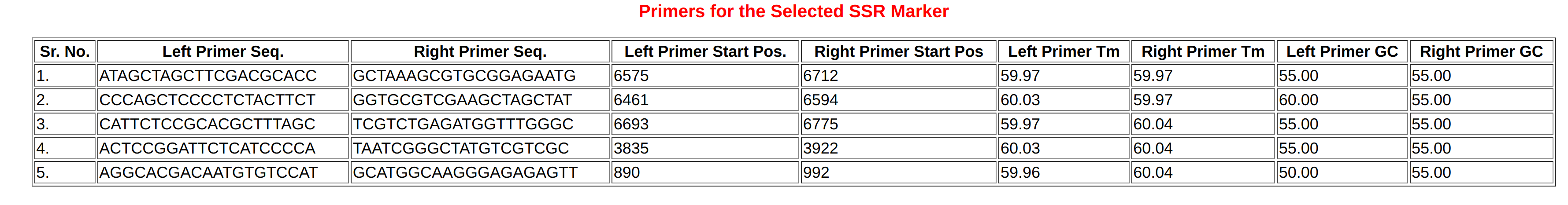
Primers are the short(18-24nt) stretch of DNA that are complementary to the target and used for gene cloning, sanger sequencing as well as expression analysis. In this database, we have provided the facility for in-silico primer designing for the query sequence. As shown in Figure-6, User can paste their target sequence (gene/cds/mrna) in the submission box in FASTA format and click on design primer button. The software automatically process the user sequence and identify most probable five set of primers along with reference values like Tm, GC percentage, start and stop position in query sequence along with primer sequences. This option is very helpful to the users wish to design primers for sequencing or expression analysis for their lab experiments.